About GEMiCCL
 Main Features
Reference
Main Features
Reference
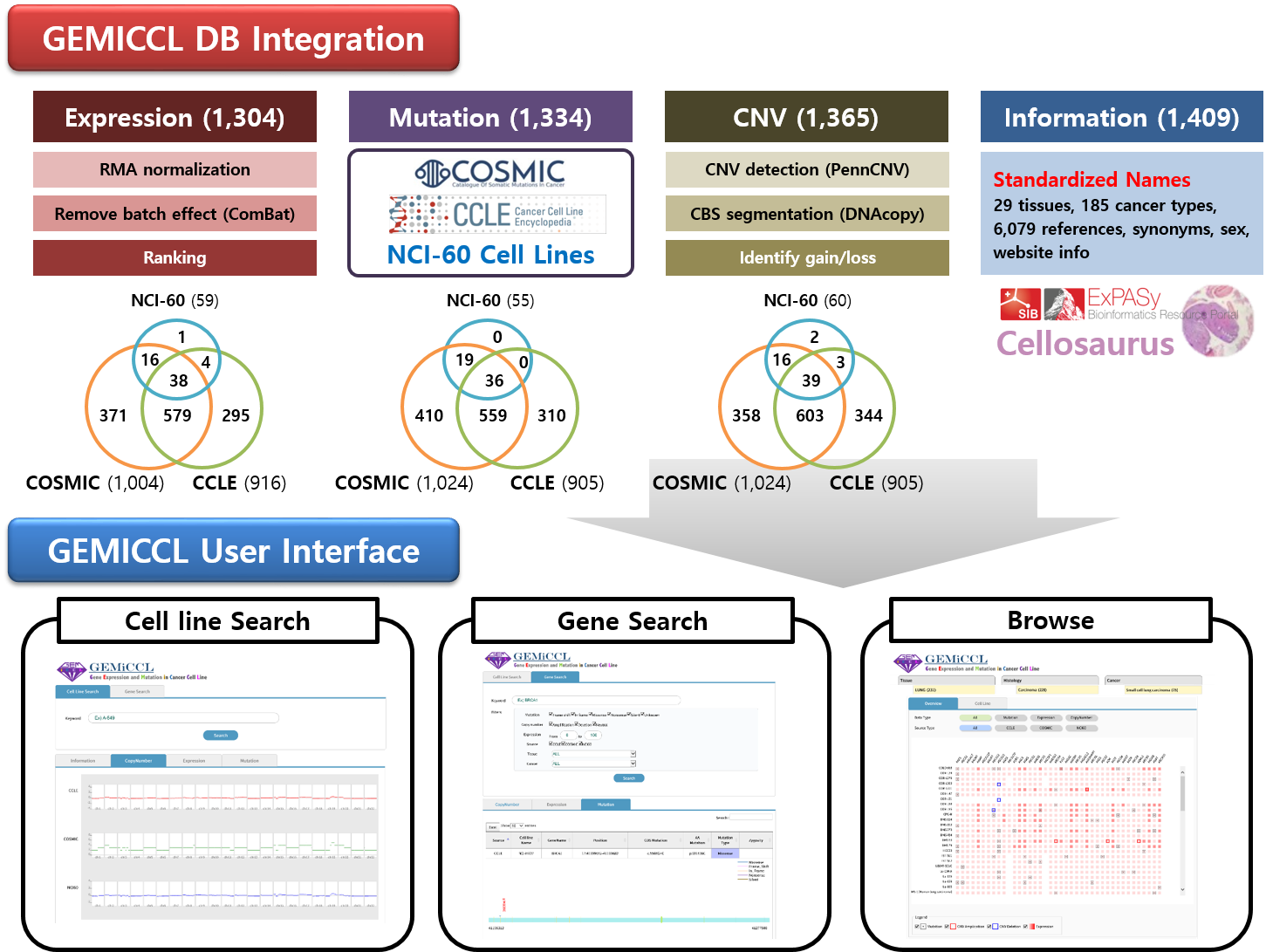
Cancer cell lines are essential components for biomedical research. However, proper choice of cell lines for experimental purposes is often difficult because genotype and/or expression data are missing or scattered in diverse resources. GEMiCCL (Gene Expression and Mutations in Cancer Cell Lines) is an online database of human cancer cell lines that provides genotype and expression information. The mutation, gene expression, and copy number variation (CNV) data were collected from three representative databases on cell lines – CCLE (Cancer Cell Line Encyclopedia), COSMIC (Catalogue of Somatic Mutations in Cancer), and NCI60. We re-processed the entire gene expression and SNP chip data and removed the batch effects due to different microarray platforms using the ComBat software.

- In total, GEMiCCL includes 1406 cell lines from 185 cancer types and 29 tissues. Gene expression, mutation, and CNV information are available for 1304, 1334, and 1365 cell lines, respectively.
- Cell line names and clinical information were standardized using Cellosaurus from ExPASy.
- Our user interface supports cell line search, gene search, browsing for specific molecular characteristics, and complex queries based on Boolean logic rules.
- We also implemented many interactive features and user-friendly visualizations including CNV plot, mutation plot, summary of molecular aberrations, and so on.
Jung I, Yu N, Jang I et al., GEMiCCL: mining genotype and expression data of cancer cell lines with elaborate visualization. submitted.


